Several types of genomic alterations can drive cancer development
Genomic alterations can drive cancer development by activating oncogenes or disrupting tumor suppressors.1
Alterations may occur as:
Gene fusions are a common class of genomic alterations4
A gene fusion is a hybrid gene formed from 2 normally separate genes, through a variety of mechanisms4:
- Chromosome rearrangement (translocations, inversions, insertions, or deletions)
- Transcription read-through
- mRNA splicing
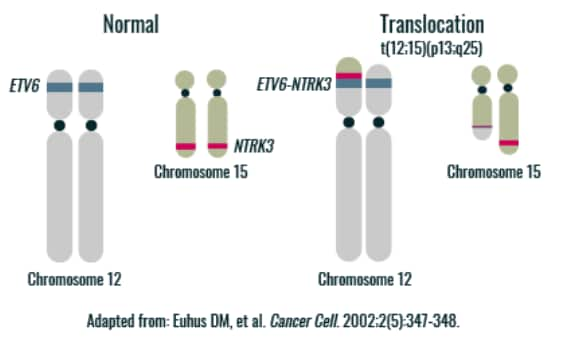
A subset of gene fusions involve NTRK1, NTRK2, and NTRK3.6
ETV6, ETS variant 6; NTRK, neurotrophic tyrosine receptor kinase; mRNA, messenger ribonucleic acid.
References
- Yates LR, Seoane J, Le Tourneau C, et al. The European Society for Medical Oncology (ESMO) Precision Medicine Glossary. Ann Oncol. 2018;29(1):30-35. Return to content
- Kumar-Sinha C, Chinnaiyan AM. Nat Biotechnol. 2018;36(1):46-60. Return to content
- Lister Hill National Center for Biomedical Communications. What kind of gene mutations are possible? Genetics Home Reference: Your Guide to Understanding Genetic Conditions. US National Library of Medicine, National Institutes of Health, US Dept of Health and Human Services. https://ghr.nlm.nih.gov/primer/mutationsanddisorders/possiblemutations. Accessed February 13, 2018. Return to content
- Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44(10):4487-4503. Return to content
- Euhus DM, Timmons CF, Tomlinson GE. ETV6-NTRK3–Trk-ing the primary event in human secretory breast cancer. Cancer Cell. 2002;2(5):347-348. Return to content
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5(1):25-34. Return to content
Active NTRK gene fusions are driver oncogenes1
UNDER NORMAL CONDITIONS, TRK RECEPTORS MEDIATE NEUROTROPHIN SIGNALING2
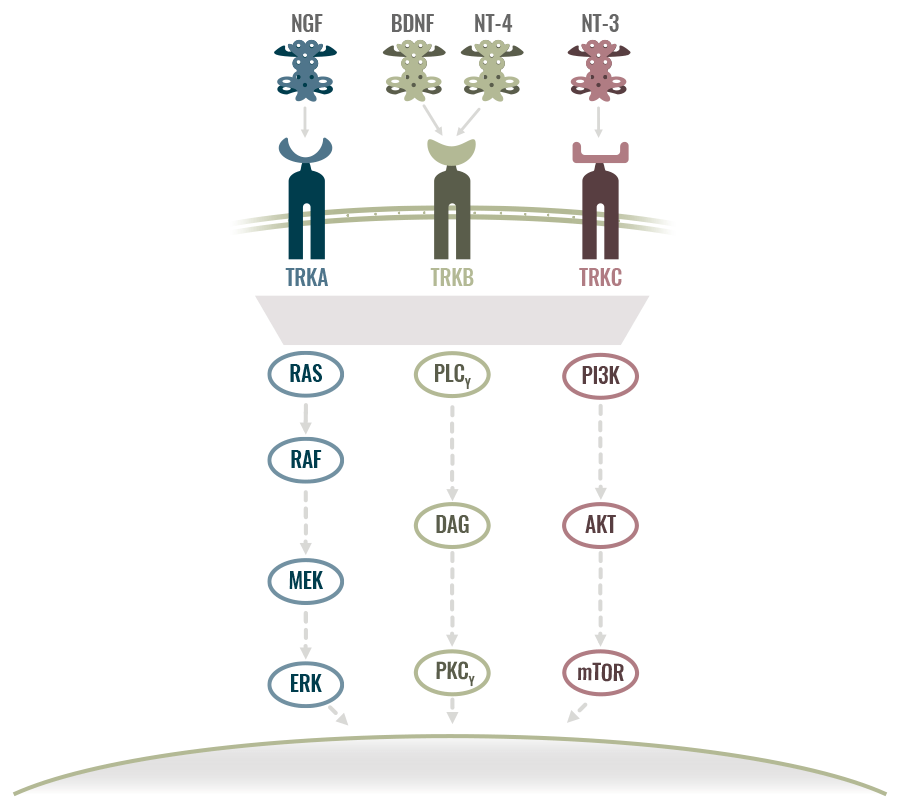
- The TRK receptor family comprises 3 transmembrane proteins referred to as TRKA, TRKB, and TRKC, encoded by the NTRK1, NTRK2, and NTRK3 genes, respectively
- TRK receptors are typically expressed in human neuronal tissue and play an essential role in the physiology, development, and function of the nervous system through activation by neurotrophins
NTRK GENE FUSIONS LEAD TO TRK FUSION PROTEINS THAT ARE ONCOGENIC DRIVERS1,2
- A subset of gene fusions involve NTRK1, NTRK2, and NTRK3 2
- Activating NTRK gene fusions couple the tyrosine kinase domain with a 5’ fusion partner resulting in a chimeric TRK protein lacking the ligand binding domain1,2
- This leads to constitutive activation of the TRK receptor kinase domain, which promotes tumor cell proliferation and survival2
BDNF, brain-derived neurotrophic factor; DNA, deoxyribonucleic acid; ERK, extracellular signal-regulated kinase; mRNA, messenger ribonucleic acid; NGF, nerve growth factor; NT, neurotrophin; NTRK, neurotrophic tyrosine receptor kinase; TRK, tropomyosin receptor kinase; Tyr, tyrosinase.
References
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5(1):25-34. 2. Amatu A, Sartore-Bianchi A, Siena S. ESMO Open. 2016;1:e000023. Return to content
NTRK gene fusions are associated with many human tumor types1,2
NTRK gene fusions are associated with a diverse range of solid tumors and hematologic malignancies1-3 and are generally mutually exclusive of other driver genomic alterations.3
NTRK GENE FUSION FREQUENCY ACROSS ADULT AND PEDIATRIC CANCERS2,5-11
PLANNING FOR
YOUR PRACTICE
Testing for NTRK gene fusions is essential to identify patients who harbor these genomic alterations12
GIST, gastrointestinal stromal tumors; NTRK, neurotrophic tyrosine receptor kinase.
References
- Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58-66. Return to content
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5(1):25-34. Return to content
- Cocco E, Scaltriti M, Drilon A. Nat Rev Clin Oncol. 2018;15(12):731-747. Return to content
- Cisowski J, Bergo MO. What makes oncogenes mutually exclusive? Small GTPases. 2017;8(3):187-192. Return to content
- Kim J, Lee Y, Cho HJ, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9(3):e91940. Return to content
- Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Comm. 2014;5:4846. Return to content
- Amatu A, Sartore-Bianchi A, Siena S. ESMO Open. 2016;1:e000023. Return to content
- Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PHB. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol. 2000;24(7):937-946. Return to content
- Rubin BP, Chen C-J, Morgan TW, et al. Congenital mesoblastic nephroma t(12; 15) is associated with ETV6-NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998;153(5):1451-1458. Return to content
- Brzeziańska E, Karbownik M, Migdalska-Sek M, Patuszak-Lewandoska D, Wloch J, Lewińska A. Molecular analysis of the RET and NTRK1 gene rearrangements in papillary thyroid carcinoma in the Polish population. Mutat Res. 2006;599(1-2):26-35. Return to content
- Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444-450. Return to content
- Kummar S, Lassen UN. Target Oncol. 2018;13(5):545-556 Return to content
NTRK gene fusion characteristics to consider for detection
NTRK gene fusions occur in numerous tumor types with multiple fusion partners.1,2 Several methods exist to detect genomic alterations; however, features unique to NTRK gene fusions must be considered when testing for NTRK gene fusions.2
- Gene fusions occur between all 3 NTRK genes (NTRK1, NTRK2, NTRK3) and fusion partners1,2
- ≥50 different fusion partners; novel partners continue to be reported1,2
- Occur across tumor types and have currently been identified in ≥20 cancers1,2
- Multiple and inconsistent genomic breakpoint locations3,4
- Large intronic regions2
- Low-complexity, GC-rich regions5,6
- TRK proteins are endogenously expressed in some tumor/tissue types1,3
GENE FUSIONS OCCUR BETWEEN ALL 3 NTRK GENES AND EACH OF SEVERAL DIFFERENT FUSION PARTNERS3
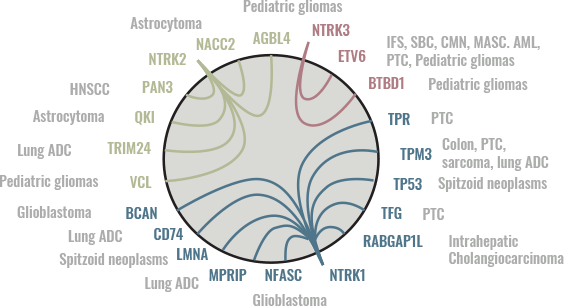
VISUALIZING NTRK GENES’ CHARACTERISTICS7
PLANNING FOR
YOUR PRACTICE
The optimal detection method for NTRK gene fusions would require no prior knowledge of fusion breakpoints and/or fusion partner, regardless of tumor type, and addresses the unique structural challenges associated with NTRK gene fusions.8
ADC, adenocarcinoma; AML, acute myelogenous leukemia; CMN, congenital mesoblastic nephroma; GC, guanine-cytosine; HNSCC, head and neck squamous cell carcinoma; IFS, infantile fibrosarcoma; MASC, mammary analogue secretory carcinoma; NTRK, neurotrophic tyrosine receptor kinase; PTC, papillary thyroid carcinoma; SBC, secretory breast cancer; TRK, tropomyosin receptor kinase.
References
- Amatu A, Sartore-Bianchi A, Siena S. ESMO Open. 2016;1:e000023. Return to content
- Kummar S, Lassen UN. Target Oncol. 2018;13(5):545-556. Return to content
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5(1):25-34. Return to content
- Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13(11):772-787. Return to content
- Abel H, Pfeifer J, Duncavage E. Translocation detection using next-generation sequencing. In: Kulkarni S, Pfeifer J, eds. Clinical Genomics. New York, NY: Elsevier/Academic Press; 2015:151-164. Return to content
- Febbo P, Champagne M. Comprehensive NTRK fusion detection, agnostic of the fusion partner, for optimal identification of rare genomic events using TruSight™ Oncology 500 [webinar]. Presented at: Drug Discovery 2019. https://www.labroots.com/virtual-event/drug-discovery-2019; February 27, 2019. Return to content
- Farago AF, Taylor MS, Doebele RC, et al. Clinicopathologic features of non–small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol. 2018. Return to content
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685-696. Return to content













