- Deviations from required preanalytical steps prior to molecular pathology can affect results2
- Biopsy sampling and preanalytical variables can affect detection quality of the different methodologies3
- Average turnaround times vary by testing methodology2
- Batching logistics vary by testing methodology4
NTRK, neurotrophic tyrosine receptor kinase.
References
- Halling KC, Wendel AJ. In situ hybridization: Principles and applications. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Biology. New York, NY: Springer Science+Business Media; 2009:109-118. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;191(3):343-365. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media; 2014:3-13. Return to content
- How turnaround time can become an issue in laboratory diagnostic testing. December 5, 2017. Diaceutics website. http://www.diaceutics.com/2017/12/05/turnaround-time-can-become-issue-laboratory-diagnostic-testing. Accessed February 23, 2019. Return to content
Different approaches to tissue acquisition are used across different tumor types
- Different volumes of tumor samples are used to support the patient’s diagnostic workup1
- Degree of biomarker testing associated with different cancer indications can vary significantly, together with volume of tumor sample used2
- For tumor types which yield small samples, such as lung cancer, on-site evaluation of tissue at biopsy can help ensure the sample is adequate for testing1
*These volumes are based on best practice recommendations and will likely vary in real-world clinical practice.
PLANNING FOR
YOUR PRACTICE
Tissue availability for NTRK gene fusion testing can vary significantly depending on tumor type.2 Thus, it is important to work together with your multidisciplinary team to ensure optimal sample availability for testing
CNB, core needle biopsy; FNA, fine needle aspiration; FNB, fine needle biopsy; GBM, glioblastoma; GIST, gastrointestinal stromal tumor; NTRK, neurotrophic tyrosine receptor kinase.
References
- Han Y, Li J. Clin Chem Lab Med. 2017;55(12):1817-1833. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341-365. Return to content
- Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol. 2012;25:S18-S30. Return to content
- Bergman RA, Afifi AK, Heidger PM. Blood. In: Bergman RA, ed. Anatomy Atlases: Atlas of Microscopic Anatomy. www.anatomyatlases.org/MicroscopicAnatomy/Section04/Section04.shtml. Accessed March 4, 2019. Return to content
- Poca MA, Martínez-Ricarte FR, Gandara DF, et al. Target location after deep cerebral biopsies using low-volume air injection in 75 patients. Results and technical note. Acta Neurochir (Wien). 2017;159(10):1939-1946. Return to content
- Hartman DJ, Slivka A, Giusto DA, Krasinskas AM. Tissue yield and diagnostic efficacy of fluoroscopic and cholangioscopic techniques to assess indeterminate biliary strictures. Clin Gastroenterol Hepatol. 2012;10:1042-1048. Return to content
- Grégoire V, Lefebvre J-L, Licitra L, Felip E. Squamous cell carcinoma of the head and neck: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21 (suppl 5):v184-v186. Return to content
- Dummer R, Hauschild A, Lindenblatt N, Pentheroudakis G, Keilhotz U; ESMO Guidelines Committee. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26 (suppl 5):v126-v132. Return to content
- Casali PG, Bielack S, Abecassis N, et al. Bone sarcomas: ESMO-PaedCan-EUROCAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21 (suppl 5): iv79-iv95. Return to content
- Pacini F, Castagna MG, Brilli L, Pentheroudakis G; ESMO Guidelines Working Group. Thyroid cancer: ESMO clinical practice guidelines for diagnosis, treatment, and follow-up. Ann Oncol. 2012;23(suppl 7):vii110-vii119. Return to content
- Rüschoff J, Hanna W, Bilous M, et al. HER2 testing in gastric cancer: a practical approach. Mod Pathol. 2012;25:637-650. Return to content
- Lee AHS, Carder P, Deb R. Guidelines for non-operative diagnostic procedures and reporting in breast cancer screening. In: Lee AHS, ed. Pathology: the Science Behind the Cure. London, UK: The Royal College of Pathologists; June 2016. Document G150. Return to content
Preanalytical steps prior to molecular pathology may affect the quality of the sample result1,2
Preanalytical step: Fixation
Areas of requirement
- Preserves tissue
- Prevents autolysis
- Preserves nucleic acids
Possible consequence of error
- Artificial mutations may be created during fixation, resulting in false positives when a highly sensitive technique (such as NGS) is used
- Decalcification of bone lesions for an excessive time can cause loss of nuclear staining and can macerate tissues, thus impacting downstream test quality
- Failure of test due to inappropriate fixation
Preanalytical step: Specimen handling
Areas of requirement
- Minimum 3-4 identifiers to match both request form and specimen
- Legible and accurate forms
Possible consequence of error
- Delay in TAT—missed window for treatment
- Sample error, patient does not receive correct treatment at the right time
Preanalytical step: Tissue processing
Areas of requirement
- To ensure all endogenous water is removed from tissue, ensuring good tissue morphology and preservation of antigen and DNA
Possible consequence of error
- DNA quality affected due to fragmentation of DNA with tissue processing
Preanalytical step: Embedding
Areas of requirement
- Specimens must be oriented correctly at embedding to ensure section includes epidermis to subcutis; this ensures pathologist can stage disease
Possible consequence of error
- Incorrect embedding of biopsy can lead to lesion being lost, inability to stage disease correctly, or inappropriate diagnosis2
FFPE samples are the standard in molecular pathology testing1-3
Preanalytical step: Processing
FFPE tissue samples
- Commonly by fixation in cold 10% NBF for the shortest possible times:
- 6-12 h for small biopsies
- 8-18 h for large surgical resections
Fresh-frozen tissues
- Embedded in optimal cutting temperature compound, frozen rapidly at −80°C, or immersed in liquid nitrogen (−190°C)
- Cut into serial frozen sections at −15°C to −25°C, using a cryostat for histological and molecular diagnoses
Preanalytical step: Advantages
FFPE tissue samples
- FFPE samples provide commendable cellular morphology and preserve the entire tumor structure for assessment of tumor cellularity
- Simple long-term storage: Can be kept in archives at room temperature for years
Fresh-frozen tissues
- Optimal for providing high-quality DNA or RNA
- More conducive to analysis of long DNA segments (>1000 bp)
Preanalytical step: Disadvantages
FFPE tissue samples
- DNA cross-linking can impact nucleic acid isolation, making it more challenging
- DNA fragmentation restricts the analysis of long DNA segments:
- Sequencing of DNA segments between 300 and 1000 bp succeeds inconsistently
- Tiling of contiguous regions to accurately identify breakpoints with sufficient coverage can be challenging
- Cytosine deamination can result in the generation of random sequence artifacts, which can potentially lead to false positives using highly sensitive techniques
- Additional pre-extraction handling procedures are required to ensure robust DNA/RNA template quality
Fresh-frozen tissues
- Fast sample processing and staining may lead to ambiguity in cell morphology (tumor vs normal tissue)
- Highly controlled storage conditions:
- Fresh surgical samples need to be kept in liquid nitrogen
- Sections that are mounted on glass slides are required to be kept at −80°C until further use
PLANNING FOR
YOUR PRACTICE
While fresh-frozen samples provide a higher-quality template, sample handling difficulties limit routine use in a real-world laboratory setting1
bp, base pairs; DNA, deoxyribonucleic acid; FFPE, formalin-fixed paraffin-embedded; NBF, neutral buffered formalin; NGS, next-generation sequencing; RNA, ribonucleic acid; TAT, turnaround time.
References
- Han Y, Li J. Clin Chem Lab Med. 2017;55(12):1817-1833. Return to content
- Rolls G. Steps to better processing and embedding. Leica Biosystems. https://www.leicabiosystems.com/pathologyleaders/steps-to-better-processing-and-embedding/. Accessed March 3, 2019. Return to content
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. Return to content
Biopsy sampling and preanalytical variables affect detection quality of the different methodologies1
FISH
- Volume of material required is linked to number of probes and reactions required
- Limited to the clinical assessment of a small number of oncogenic markers6
PLANNING FOR
YOUR PRACTICE
Samples should be carefully reviewed by pathologists to identify tumor-rich areas and ensure biopsy sample contains high tumor content that can support all methodologies1
DNA, deoxyribonucleic acid; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NGS, next-generation sequencing; RNA, ribonucleic acid; RT-PCR, reverse-transcription polymerase chain reaction.
References
- Han Y, Li J. Clin Chem Lab Med. 2017;55(12):1817-1833. Return to content
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685-696. Return to content
- Serrati S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355-7365. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341-365. Return to content
- Fitzgibbons PL, Cooper K. Immunohistochemistry of biomarkers. In Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. New York, NY: Springer Science+Business Media; 2009:133-137. Return to content
- Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol. 2018;31(3):463-473. Return to content
- Olsen JL. Polymerase chain reaction. In: Vohr HW, ed. Encyclopedia of Immunotoxicology. Berlin, Germany; Springer Verlag 2016:715-720. Return to content
Average turnaround times* vary by methodology used1
Laboratory TATs associated with common gene fusion methodologies
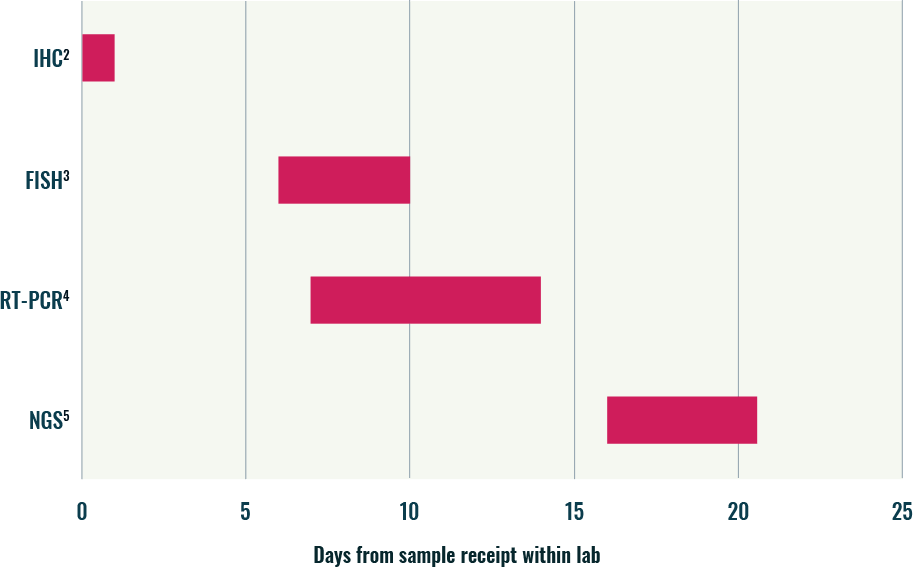
*This may be due to individual batching considerations within the laboratory.
- When comparing average TAT associated with different methodologies, NGS typically takes the longest time6
- This is largely due to the complexities associated with the test set up and bioinformatics analysis6
- IHC, FISH, and RT-PCR have faster turnaround times, reflecting their incorporation within established laboratory workflows supporting time efficiencies6
Recognizing potential issues that can affect testing turnaround time
Sequential reflex testing
- May be required or necessary to confirm NTRK gene fusion–positive results determined by one method (ie, FISH/IHC) that requires another molecular method (ie, NGS)7
Sample handling and preanalytical errors
- General sample shipping and storage requirements may impact transit time8
- Inappropriate handling of primary samples may result in suboptimal DNA quality, thus requiring repeat testing, which will extend TAT6,8
Sample batching
- Labs may test once or twice a week. This enables a cost-efficient service but results in delays of TAT (eg, waiting until you have enough samples to build a library to optimize cost/run because reagents are usually one-time use only)9
Methodology
- Testing method used will dictate turnaround time (ie, TAT for IHC is generally shorter than FISH, IHC/FISH shorter than NGS, etc)2,3,5,10
DNA-/RNA-based NGS
- Factors specific to NGS—such as DNA/RNA extraction, library prep, sequencing time, data analysis/annotation, and reporting—all contribute to relatively longer NGS TATs, which can range from 10-30 days11
DNA, deoxyribonucleic acid; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NGS, next-generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; RNA, ribonucleic acid; RT-PCR, reverse-transcription polymerase chain reaction; TAT, turnaround time.
References
- How turnaround time can become an issue in laboratory diagnostic testing. December 5, 2017. Diaceutics website. http://www.diaceutics.com/2017/12/05/turnaround-time-can-become-issue-laboratory-diagnostic-testing. Accessed February 23, 2019. Return to content
- Rudzinski ER, Lockwood CM, Stohr BA, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol.> 2018;42(7):927-935. Return to content
- Indiana University Department of Medical and Molecular Genetics. Specimen criteria for clients. http://geneticslab.medicine.iu.edu/Files/Specimen%20Criteria%20for%20Clients%20V07182018.pdf. Accessed March 1, 2019. Return to content
- Indiana University Department of Medical and Molecular Genetics. Specimen criteria for clients. http://geneticslab.medicine.iu.edu/Files/Specimen%20Criteria%20for%20Clients%20V07182018.pdf. Accessed March 1, 2019. Return to content
- Devarakonda S. Expert highlights benefits of next-generation sequencing for NSCLC. http://www.targetedonc.com/news/expert-highlights-benefits-of-nextgeneration-sequencing-for-nsclc. Accessed March 1, 2019. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media; 2014:3-13. Return to content
- Farago AF, Taylor MS, Doebele RC, et al. Clinicopathologic features of non–small-cell lung cancer harbouring an NTRK gene fusion. JCO Precis Oncol. 2018. Return to content
- Han Y, Li J. Clin Chem Lab Med. 2017;55(12):1817-1833. Return to content
- Batch vs continuous flow specimen processing. Lab CE website. https://www.labce.com/spg1837894_batch_versus_continuous_flow_specimen_processing.aspx. Accessed February 26, 2019. Return to content
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333-351. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;191(3):341-365. Return to content
- Serrati S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355-7365. Return to content
- Kummar S, Lassen UN. Target Oncol. 2018:13(5):545-556. Return to content
- Jang JS, Wang X, Vedell PT, et al. Custom gene capture and next-generation sequencing to resolve discordant ALK status by FISH and IHC in lung adenocarcinoma. J Thorac Oncol. 2016;11(11):1891-1900. Return to content
- Gounder MM, Ali SM, Robinson V, et al. Impact of next-generation sequencing (NGS) on diagnostic and therapeutic options in soft-tissue and bone sarcoma. J Clin Oncol. 2017;35(suppl). Abstract 11001. Return to content
Laboratory considerations associated with common methodologies, which aid the implementation of efficient workflows
*Depends on number of reactions and fusions to detect.5
FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NGS, next-generation sequencing; RT-PCR, reverse-transcription polymerase chain reaction.
References
- Next Generation Sequencing Implementation Guide. Silver Spring, MD: Association of Public Health Laboratories; 2016. Return to content
- Serratì S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355-7365. Return to content
- Chen ZE, Lin F. Overview of predictive biomarkers and integration of IHC into molecular pathology. In: Lin F, Prichard J, eds. Handbook of Practical Immunohistochemistry: Frequently Asked Questions. New York, NY: Springer Science+Business Media; 2015:91-104. Return to content
- Hu L, Ru K, Zhang L, et al. Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. Biomark Res. 2014;2(1):1-13. Return to content
- Hu L, Ru K, Zhang L, et al. Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. Biomark Res. 2014;2(1):1-13. Return to content
- Darawi MN, Ai-Vyrn C, Ramasamy K, et al. Allele-specific polymerase chain reaction for the detection of Alzheimer’s disease-related single nucleotide polymorphisms. BMC Med Genet. 2013;14:27. Return to content
- Peter M, Gilbert E, Delattre O. A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest. 2001;81(6):905-912. Return to content





